Hi! This last post is about the last three weeks of my time as an Amherst College SURF researcher and, as such, it will be a long one, so be sure to read ‘til the end!
Since my last post, I have finished my Illustrator and Igor training and have now moved onto performing research (woo!). This has meant developing a whole new set of skills as well as incorporating what I have already learned from my training.
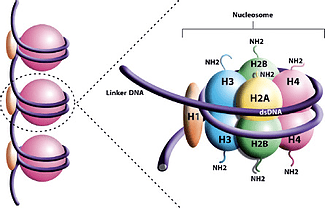
As a member of the Histone Research Group in the Carter Lab, the focus of my research is determining the steps that lead to the creation of a Nucleosome Core Particle (NCP), a structure in our genome that consists of our DNA wrapped about 1.7 times around eight histone proteins all held together with a special “H1” histone. From what we know, the eight histone proteins come together as pairs of two, or dimers, until an octamer is formed.
In order to collect data on how the NCP is created, we use a combination of Gwyddion, AFM images, Microsoft Excel, and Igor. While Igor and AFM may sound familiar from my last blog post, Gwyddion has been unheard of up until now. It is the program used to process the images created from the AFM, as well as for the collection of data from our slides.
The data collection process includes going through each individual singlet — or DNA strand with or without histone — and measuring values such as DNA length, histone color, and the number of histones in the singlet. Each of these data values is recorded in Excel files, which are then shared with the rest of the group via DropBox.
Of course, I am able to explain it clearly now but I was totally lost on all of this when I first began! I had no idea how to even measure a DNA strand, but, once again, the importance of self-advocacy came into play just as it had when I was learning Igor and so I always asked every question I had.
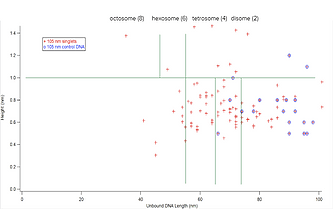
In preparation for a video I was to make for the SURF program, I was tasked with creating a graph in Igor that incorporates data from105 nm DNA slides — specifically, a graph plotting histone height as a function of unbound DNA length.
The rationale behind the data used for the graph is as follows: as more histone proteins are bound together, there would be more wrapping of the DNA, thus resulting in a shorter length of unbound DNA length. Thus, what we would expect to see in our graph is a negative correlation.
Despite my training having been done, my programming work with Igor was not — although, I have to say, this program was a lot more enjoyable to work with as I now had some programming experience under my belt and it also incorporated Illustrator. This time, I had to program a game of hangman — which, turned out to be a lot more complicated than it may sound at first, but with the help of my fellow SURF pals and my professor it was possible!
Throughout all of this, I was still finding ways to bridge the gap between the SURF students in addition to the weekly Book Club I was participating in, and our editor-in-chief and I were able to do just that with a Kahoot-style Trivia Night! Not only did we get to see some new and familiar faces, we were also able to give five lucky contestants a very cute axolotl plush!
The culmination of my work is in a three-minute Flash Talk video, created for the SURF program. I am unable to share it via this blog post, but soon it will be available on the ASN website, so be on the lookout.
But just because SURF is over now does not mean that my work in Amherst’s STEM community is over yet!
Right before publishing my last blog post, I became a member of Amherst College’s Association for Women In Science (AWIS) e-board and have begun work on organizing a virtual panel scheduled for Spring 2021. Additionally, I will be participating in a SURF presentation on behalf of the Departments of Physics and Astronomy in early September, so stay tuned for that!

Do not forget, I will always be available at jidrovo23@amherst.edu if you have any questions!


You must be logged in to post a comment.